Mutations in SCNM1 cause orofaciodigital syndrome due to minor intron splicing defects affecting primary cilia
- PMID: 36084634
- PMCID: PMC9606384
- DOI: 10.1016/j.ajhg.2022.08.009
Abstract
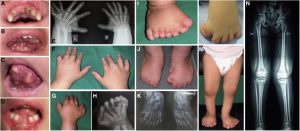
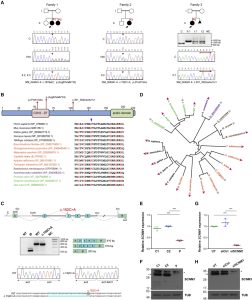
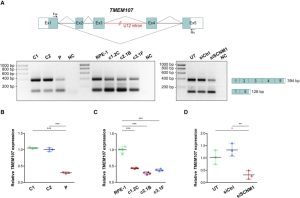
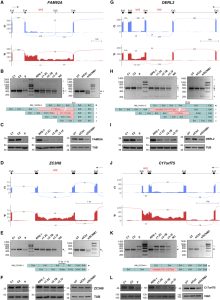
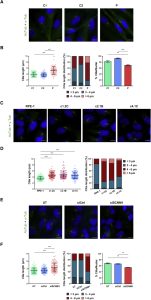
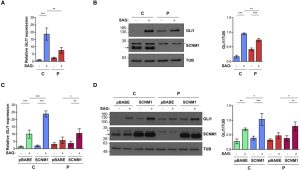
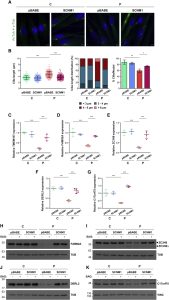
Orofaciodigital syndrome (OFD) is a genetically heterogeneous ciliopathy characterized by anomalies of the oral cavity, face, and digits. We describe individuals with OFD from three unrelated families having bi-allelic loss-of-function variants in SCNM1 as the cause of their condition. SCNM1 encodes a protein recently shown to be a component of the human minor spliceosome. However, so far the effect of loss of SCNM1 function on human cells had not been assessed. Using a comparative transcriptome analysis between fibroblasts derived from an OFD-affected individual harboring SCNM1 mutations and control fibroblasts, we identified a set of genes with defective minor intron (U12) processing in the fibroblasts of the affected subject. These results were reproduced in SCNM1 knockout hTERT RPE-1 (RPE-1) cells engineered by CRISPR-Cas9-mediated editing and in SCNM1 siRNA-treated RPE-1 cultures. Notably, expression of TMEM107 and FAM92A encoding primary cilia and basal body proteins, respectively, and that of DERL2, ZC3H8, and C17orf75, were severely reduced in SCNM1-deficient cells. Primary fibroblasts containing SCNM1 mutations, as well as SCNM1 knockout and SCNM1 knockdown RPE-1 cells, were also found with abnormally elongated cilia. Conversely, cilia length and expression of SCNM1-regulated genes were restored in SCNM1-deficient fibroblasts following reintroduction of SCNM1 via retroviral delivery. Additionally, functional analysis in SCNM1-retrotransduced fibroblasts showed that SCNM1 is a positive mediator of Hedgehog (Hh) signaling. Our findings demonstrate that defective U12 intron splicing can lead to a typical ciliopathy such as OFD and reveal that primary cilia length and Hh signaling are regulated by the minor spliceosome through SCNM1 activity.
Keywords: SCNM1; U12 introns; ciliopathy; hedgehog signaling; minor spliceosome; orofaciodigital syndrome; primary cilia.
Copyright © 2022 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Similar articles
-
Genet Med. 2024 Apr;26(4):101059. doi: 10.1016/j.gim.2023.101059. Epub 2023 Dec 27.PMID: 38158857
-
Plant J. 2014 Jun;78(5):799-810. doi: 10.1111/tpj.12498. Epub 2014 Apr 15.PMID: 24606192
-
Hum Mutat. 2016 Feb;37(2):155-9. doi: 10.1002/humu.22925. Epub 2015 Nov 23.PMID: 26518474
-
Dysregulated minor intron splicing in cancer.
Cancer Sci. 2022 Sep;113(9):2934-2942. doi: 10.1111/cas.15476. Epub 2022 Jul 11.PMID: 35766428Free PMC article.Review. -
The role of splicing factors in retinitis pigmentosa: links to cilia.
Biochem Soc Trans. 2021 Jun 30;49(3):1221-1231. doi: 10.1042/BST20200798.PMID: 34060618Review.
Cited by
-
Ann Surg Oncol. 2025 Jun;32(6):4508-4519. doi: 10.1245/s10434-025-17108-z. Epub 2025 Apr 2.PMID: 40172715
-
Connecting genotype and phenotype in minor spliceosome diseases.
RNA. 2025 Feb 19;31(3):284-299. doi: 10.1261/rna.080337.124.PMID: 39761998Free PMC article.Review. -
Role of U11/U12 minor spliceosome gene ZCRB1 in Ciliogenesis and WNT Signaling.
bioRxiv [Preprint]. 2024 Aug 10:2024.08.09.607392. doi: 10.1101/2024.08.09.607392.PMID: 39149385Free PMC article.Preprint. -
Hum Mol Genet. 2023 Sep 5;32(18):2822-2831. doi: 10.1093/hmg/ddad109.PMID: 37384395Free PMC article.
-
Nat Commun. 2025 May 22;16(1):4741. doi: 10.1038/s41467-025-59998-3.PMID: 40399278Free PMC article.
References
- El Marabti E., Malek J., Younis I. Minor intron splicing from basic science to disease. Int. J. Mol. Sci. 2021;22:6062. – PMC – PubMed
- Verma B., Akinyi M.V., Norppa A.J., Frilander M.J. Minor spliceosome and disease. Semin. Cell Dev. Biol. 2018;79:103–112. – PubMed
- Sharp P.A., Burge C.B. Classification of Introns: U2-Type or U12-Type. Cell. 1997;91:875–879. – PubMed
- Turunen J.J., Niemela E.H., Verma B., Frilander M.J. Vol. 4. Wiley Interdiscip Rev RNA; 2013. The Significant Other: Splicing by the Minor Spliceosome; pp. 61–76. – PMC – PubMed
- Baumgartner M., Drake K., Kanadia R.N. An integrated model of minor intron emergence and conservation. Front. Genet. 2019;10:1113. – PMC – PubMed
- Bartschat S., Samuelsson T. U12 type introns were lost at multiple occasions during evolution. BMC Genom. 2010;11:106. – PMC – PubMed
- Olthof A.M., Hyatt K.C., Kanadia R.N. Minor intron splicing revisited: identification of new minor intron-containing genes and tissue-dependent retention and alternative splicing of minor introns. BMC Genom. 2019;20:686. – PMC – PubMed
- Montzka K.A., Steitz J.A. Additional low-abundance human small nuclear ribonucleoproteins: U11, U12, etc. Proc. Natl. Acad. Sci. USA. 1988;85:8885–8889. – PMC – PubMed
- Patel A.A., McCarthy M., Steitz J.A. The splicing of U12-type introns can be a rate-limiting step in gene expression. EMBO J. 2002;21:3804–3815. – PMC – PubMed
- Xing C., Kanchwala M., Rios J.J., Hyatt T., Wang R.C., Tran A., Dougherty I., Tovar-Garza A., Purnadi C., Kumar M.G., et al. Biallelic variants in RNU12 cause CDAGS syndrome. Hum. Mutat. 2021;42:1042–1052. – PubMed
- Olthof A.M., Rasmussen J.S., Campeau P.M., Kanadia R.N. Disrupted minor intron splicing is prevalent in Mendelian disorders. Mol. Genet. Genomic Med. 2020;8:e1374. – PMC – PubMed
- Garcia-Gonzalo F.R., Reiter J.F. Open sesame: how transition fibers and the transition zone control ciliary composition. Cold Spring Harb. Perspect. Biol. 2017;9:a028134. – PMC –PubMed
- Nachury M.V., Mick D.U. Establishing and regulating the composition of cilia for signal transduction. Nat. Rev. Mol. Cell Biol. 2019;20:389–405. – PMC – PubMed
- Lechtreck K.F. IFT–cargo interactions and protein transport in cilia. Trends Biochem. Sci. 2015;40:765–778. – PMC – PubMed
- Bangs F., Anderson K.V. Primary cilia and mammalian hedgehog signaling. Cold Spring Harb. Perspect. Biol. 2017;9:a028175. – PMC – PubMed
- Kong J.H., Siebold C., Rohatgi R. Biochemical mechanisms of vertebrate hedgehog signaling. Development. 2019;146 – PMC – PubMed
- Liu A., Wang B., Niswander L.A. Mouse intraflagellar transport proteins regulate both the activator and repressor functions of Gli transcription factors. Development. 2005;132:3103–3111. – PubMed
- Huangfu D., Anderson K.V. Cilia and Hedgehog responsiveness in the mouse. Proc. Natl. Acad. Sci. USA. 2005;102:11325–11330. – PMC – PubMed
- Sreekumar V., Norris D.P. Cilia and development. Curr. Opin. Genet. Dev. 2019;56:15–21. –PubMed
- Franco B., Thauvin-Robinet C. Update on oral-facial-digital syndromes (OFDS) Cilia. 2016;5:12. – PMC – PubMed
- Gurrieri F., Franco B., Toriello H., Neri G. Oral-facial-digital syndromes: review and diagnostic guidelines. Am. J. Med. Genet. 2007;143A:3314–3323. – PubMed
- Bruel A.L., Franco B., Duffourd Y., Thevenon J., Jego L., Lopez E., Deleuze J.F., Doummar D., Giles R.H., Johnson C.A., et al. Fifteen years of research on oral-facial-digital syndromes: from 1 to 16 causal genes. J. Med. Genet. 2017;54:371–380. – PMC – PubMed
- Shaheen R., Jiang N., Alzahrani F., Ewida N., Al-Sheddi T., Alobeid E., Musaev D., Stanley V., Hashem M., Ibrahim N., et al. Bi-allelic mutations in FAM149B1 cause abnormal primary cilium and a range of ciliopathy phenotypes in humans. Am. J. Hum. Genet. 2019;104:731–737. –PMC – PubMed
- Yamada M., Uehara T., Suzuki H., Takenouchi T., Fukushima H., Morisada N., Tominaga K., Onoda M., Kosaki K. IFT172 as the 19th gene causative of oral-facial-digital syndrome. Am. J. Med. Genet. 2019;179:2510–2513. – PubMed
- Bai R., Wan R., Wang L., Xu K., Zhang Q., Lei J., Shi Y. Structure of the activated human minor spliceosome. Science. 2021;371:eabg0879. – PubMed
- Palencia-Campos A., Ullah A., Nevado J., Yildirim R., Unal E., Ciorraga M., Barruz P., Chico L., Piceci-Sparascio F., Guida V., et al. GLI1 inactivation is associated with developmental phenotypes overlapping with Ellis-van Creveld syndrome. Hum. Mol. Genet. 2017;26:4556–4571. – PubMed
- Estañ M.C., Fernández-Núñez E., Zaki M.S., Esteban M.I., Donkervoort S., Hawkins C., Caparros-Martin J.A., Saade D., Hu Y., Bolduc V., et al. Recessive mutations in muscle-specific isoforms of FXR1 cause congenital multi-minicore myopathy. Nat. Commun. 2019;10:797. – PMC – PubMed
- 1000 Genomes Project Consortium. Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., et al. A global reference for human genetic variation. Nature. 2015;526:68–74. – PMC – PubMed
- Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., et al. The mutational constraint spectrum quantified from variation in 141, 456 humans. Nature. 2020;581:434–443. – PMC – PubMed
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. –PMC – PubMed
- Oghabian A., Greco D., Frilander M.J. IntEREst: intron-exon retention estimator. BMC Bioinf. 2018;19:130. – PMC – PubMed
- Garrido-Martín D., Palumbo E., Guigó R., Breschi A. ggsashimi: Sashimi plot revised for browser- and annotation-independent splicing visualization. PLoS Comput. Biol. 2018;14:e1006360. – PMC – PubMed
- Caparrós-Martín J.A., Valencia M., Reytor E., Pacheco M., Fernandez M., Perez-Aytes A., Gean E., Lapunzina P., Peters H., Goodship J.A., Ruiz-Perez V.L. The ciliary Evc/Evc2 complex interacts with Smo and controls Hedgehog pathway activity in chondrocytes by regulating Sufu/Gli3 dissociation and Gli3 trafficking in primary cilia. Hum. Mol. Genet. 2013;22:124–139. – PubMed
- Edgar R.C. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinf. 2004;5:113. – PMC – PubMed
- Okonechnikov K., Golosova O., Fursov M., UGENE team Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics. 2012;28:1166–1167. – PubMed
- Letunic I., Bork P. Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49:W293–W296. – PMC – PubMed
- Nagy E., Maquat L.E. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem. Sci. 1998;23:198–199. – PubMed
- Reese M.G., Eeckman F.H., Kulp D., Haussler D. Improved splice site detection in Genie. J. Comput. Biol. 1997;4:311–323. – PubMed
- Kohany O., Gentles A.J., Hankus L., Jurka J. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinf. 2006;7:474. –PMC – PubMed
- Deininger P. Alu elements: know the SINEs. Genome Biol. 2011;12:236. – PMC – PubMed
- Bujakowska K.M., White J., Place E., Consugar M., Comander J. Efficient in silico identification of a common insertion in the MAK gene which causes retinitis pigmentosa. PLoS One. 2015;10:e0142614. – PMC – PubMed
- Torene R.I., Galens K., Liu S., Arvai K., Borroto C., Scuffins J., Zhang Z., Friedman B., Sroka H., Heeley J., et al. Mobile element insertion detection in 89, 874 clinical exomes. Genet. Med. 2020;22:974–978. – PMC – PubMed
- Burge C.B., Padgett R.A., Sharp P.A. Evolutionary fates and origins of U12-type introns. Mol. Cell. 1998;2:773–785. – PubMed
- Dávila López M., Rosenblad M.A., Samuelsson T. Computational screen for spliceosomal RNA genes aids in defining the phylogenetic distribution of major and minor spliceosomal components. Nucleic Acids Res. 2008;36:3001–3010. – PMC – PubMed
- Howell V.M., de Haan G., Bergren S., Jones J.M., Culiat C.T., Michaud E.J., Frankel W.N., Meisler M.H. A targeted deleterious allele of the splicing factor SCNM1 in the mouse. Genetics. 2008;180:1419–1427. – PMC – PubMed
- Vogel C., Abreu R.d.S., Ko D., Le S.Y., Shapiro B.A., Burns S.C., Sandhu D., Boutz D.R., Marcotte E.M., Penalva L.O. Sequence signatures and mRNA concentration can explain two-thirds of protein abundance variation in a human cell line. Mol. Syst. Biol. 2010;6:400. – PMC –PubMed
- Roberson E.C., Dowdle W.E., Ozanturk A., Garcia-Gonzalo F.R., Li C., Halbritter J., Elkhartoufi N., Porath J.D., Cope H., Ashley-Koch A., et al. TMEM231, mutated in orofaciodigital and Meckel syndromes, organizes the ciliary transition zone. J. Cell Biol. 2015;209:129–142. –PMC – PubMed
- Shylo N.A., Christopher K.J., Iglesias A., Daluiski A., Weatherbee S.D. TMEM107 is a critical regulator of ciliary protein composition and is mutated in orofaciodigital syndrome. Hum. Mutat. 2016;37:155–159. – PubMed
- Wang C., Li J., Meng Q., Wang B. Three Tctn proteins are functionally conserved in the regulation of neural tube patterning and Gli3 processing but not ciliogenesis and Hedgehog signaling in the mouse. Dev. Biol. 2017;430:156–165. – PMC – PubMed
- Verma P.K., El-Harouni A.A. Review of literature: genes related to postaxial polydactyly. Front. Pediatr. 2015;3:8. – PMC – PubMed
- Buchner D.A., Trudeau M., Meisler M.H. SCNM1, a putative RNA splicing factor that modifies disease severity in mice. Science. 2003;301:967–969. – PubMed
- Sprunger L.K., Escayg A., Tallaksen-Greene S., Albin R.L., Meisler M.H. Dystonia associated with mutation of the neuronal sodium channel Scn8a and identification of the modifier locus Scnm1 on mouse chromosome 3. Hum. Mol. Genet. 1999;8:471–479. – PubMed
- Meisler M.H. SCN8A encephalopathy: mechanisms and models. Epilepsia. 2019;60(Suppl 3):S86–S91. – PMC – PubMed
- Argente J., Flores R., Gutiérrez-Arumí A., Verma B., Martos-Moreno G.Á., Cuscó I., Oghabian A., Chowen J.A., Frilander M.J., Pérez-Jurado L.A. Defective minor spliceosome mRNA processing results in isolated familial growth hormone deficiency. EMBO Mol. Med. 2014;6:299–306. – PMC – PubMed
- Olthof A.M., White A.K., Mieruszynski S., Doggett K., Lee M.F., Chakroun A., Abdel Aleem A.K., Rousseau J., Magnani C., Roifman C.M., et al. Disruption of exon-bridging interactions between the minor and major spliceosomes results in alternative splicing around minor introns. Nucleic Acids Res. 2021;49:3524–3545. – PMC – PubMed
- Lambacher N.J., Bruel A.L., van Dam T.J.P., Szymańska K., Slaats G.G., Kuhns S., McManus G.J., Kennedy J.E., Gaff K., Wu K.M., et al. TMEM107 recruits ciliopathy proteins to subdomains of the ciliary transition zone and causes Joubert syndrome. Nat. Cell Biol. 2016;18:122–131. – PMC – PubMed
- Shaheen R., Almoisheer A., Faqeih E., Babay Z., Monies D., Tassan N., Abouelhoda M., Kurdi W., Al Mardawi E., Khalil M.M.I., et al. Identification of a novel MKS locus defined by TMEM107 mutation. Hum. Mol. Genet. 2015;24:5211–5218. – PubMed
- Christopher K.J., Wang B., Kong Y., Weatherbee S.D. Forward genetics uncovers Transmembrane protein 107 as a novel factor required for ciliogenesis and Sonic hedgehog signaling. Dev. Biol. 2012;368:382–392. – PMC – PubMed
- Li F.Q., Chen X., Fisher C., Siller S.S., Zelikman K., Kuriyama R., Takemaru K.I. BAR domain-containing FAM92 proteins interact with chibby1 to facilitate ciliogenesis. Mol. Cell Biol. 2016;36:2668–2680. – PMC – PubMed
- Schrauwen I., Giese A.P., Aziz A., Lafont D.T., Chakchouk I., Santos-Cortez R.L.P., Lee K., Acharya A., Khan F.S., Ullah A., et al. FAM92A underlies nonsyndromic postaxial polydactyly in humans and an abnormal limb and digit skeletal phenotype in mice. J. Bone Miner. Res. 2019;34:375–386. – PMC – PubMed
- Navarro Negredo P., Edgar J.R., Manna P.T., Antrobus R., Robinson M.S. The WDR11 complex facilitates the tethering of AP-1-derived vesicles. Nat. Commun. 2018;9:596. – PMC – PubMed
- Kim Y.J., Osborn D.P., Lee J.Y., Araki M., Araki K., Mohun T., Känsäkoski J., Brandstack N., Kim H.T., Miralles F., et al. WDR11-mediated Hedgehog signalling defects underlie a new ciliopathy related to Kallmann syndrome. EMBO Rep. 2018;19:269–289. – PMC – PubMed
- Egloff S., Vitali P., Tellier M., Raffel R., Murphy S., Kiss T. The 7SK snRNP associates with the little elongation complex to promote snRNA gene expression. EMBO J. 2017;36:934–948. –PMC – PubMed
- Schmidt J.A., Danielson K.G., Duffner E.R., Radecki S.G., Walker G.T., Shelton A., Wang T., Knepper J.E. Regulation of the oncogenic phenotype by the nuclear body protein ZC3H8. BMC Cancer. 2018;18:759. – PMC – PubMed
- Dickinson M.E., Flenniken A.M., Ji X., Teboul L., Wong M.D., White J.K., Meehan T.F., Weninger W.J., Westerberg H., Adissu H., et al. High-throughput discovery of novel developmental phenotypes. Nature. 2016;537:508–514. – PMC – PubMed
- Oda Y., Okada T., Yoshida H., Kaufman R.J., Nagata K., Mori K. Derlin-2 and Derlin-3 are regulated by the mammalian unfolded protein response and are required for ER-associated degradation. J. Cell Biol. 2006;172:383–393. – PMC – PubMed
- Huang C.H., Hsiao H.T., Chu Y.R., Ye Y., Chen X. Derlin2 protein facilitates HRD1-mediated retro-translocation of sonic hedgehog at the endoplasmic reticulum. J. Biol. Chem. 2013;288:25330–25339. – PMC – PubMed
- Dougan S.K., Hu C.C.A., Paquet M.E., Greenblatt M.B., Kim J., Lilley B.N., Watson N., Ploegh H.L. Derlin-2-deficient mice reveal an essential role for protein dislocation in chondrocytes. Mol. Cell Biol. 2011;31:1145–1159. – PMC – PubMed
- Funabashi T., Katoh Y., Okazaki M., Sugawa M., Nakayama K. Interaction of heterotrimeric kinesin-II with IFT-B-connecting tetramer is crucial for ciliogenesis. J. Cell Biol. 2018;217:2867–2876. – PMC – PubMed
- Nishijima Y., Hagiya Y., Kubo T., Takei R., Katoh Y., Nakayama K. RABL2 interacts with the intraflagellar transport-B complex and CEP19 and participates in ciliary assembly. Mol. Biol. Cell. 2017;28:1652–1666. – PMC – PubMed
- Kanie T., Abbott K.L., Mooney N.A., Plowey E.D., Demeter J., Jackson P.K. The CEP19-RABL2 GTPase complex binds IFT-B to initiate intraflagellar transport at the ciliary base. Dev. Cell. 2017;42:22–36.e12. – PMC – PubMed
- Grosso A.R., Gomes A.Q., Barbosa-Morais N.L., Caldeira S., Thorne N.P., Grech G., von Lindern M., Carmo-Fonseca M. Tissue-specific splicing factor gene expression signatures. Nucleic Acids Res. 2008;36:4823–4832. – PMC – PubMed
- Avasthi P., Marshall W.F. Stages of ciliogenesis and regulation of ciliary length. Differentiation. 2012;83:S30–S42. – PMC – PubMed
- Cantagrel V., Silhavy J.L., Bielas S.L., Swistun D., Marsh S.E., Bertrand J.Y., Audollent S., Attié-Bitach T., Holden K.R., Dobyns W.B., et al. Mutations in the cilia gene ARL13B lead to the classical form of Joubert syndrome. Am. J. Hum. Genet. 2008;83:170–179. – PMC – PubMed